CF520C DNA Alignment
Description
Vasya became interested in bioinformatics. He's going to write an article about similar cyclic DNA sequences, so he invented a new method for determining the similarity of cyclic sequences.
Let's assume that strings $ s $ and $ t $ have the same length $ n $ , then the function $ h(s,t) $ is defined as the number of positions in which the respective symbols of $ s $ and $ t $ are the same. Function $ h(s,t) $ can be used to define the function of Vasya distance $ ρ(s,t) $ :
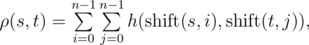 where  is obtained from string $ s $ , by applying left circular shift $ i $ times. For example, $ ρ("AGC","CGT")= $ $ h("AGC","CGT")+h("AGC","GTC")+h("AGC","TCG")+ $ $ h("GCA","CGT")+h("GCA","GTC")+h("GCA","TCG")+ $ $ h("CAG","CGT")+h("CAG","GTC")+h("CAG","TCG")= $ $ 1+1+0+0+1+1+1+0+1=6 $ Vasya found a string $ s $ of length $ n $ on the Internet. Now he wants to count how many strings $ t $ there are such that the Vasya distance from the string $ s $ attains maximum possible value. Formally speaking, $ t $ must satisfy the equation: .
Vasya could not try all possible strings to find an answer, so he needs your help. As the answer may be very large, count the number of such strings modulo $ 10^{9}+7 $ .
Input Format
The first line of the input contains a single integer $ n $ ( $ 1
Output Format
Print a single number — the answer modulo $ 10^{9}+7 $ .
Explanation/Hint
Please note that if for two distinct strings $ t_{1} $ and $ t_{2} $ values $ ρ(s,t_{1}) $ и $ ρ(s,t_{2}) $ are maximum among all possible $ t $ , then both strings must be taken into account in the answer even if one of them can be obtained by a circular shift of another one.
In the first sample, there is $ ρ("C","C")=1 $ , for the remaining strings $ t $ of length 1 the value of $ ρ(s,t) $ is 0.
In the second sample, $ ρ("AG","AG")=ρ("AG","GA")=ρ("AG","AA")=ρ("AG","GG")=4 $ .
In the third sample, $ ρ("TTT","TTT")=27 $